
Whole Genome Sequencing Part 2: Using WGS Extract
Posted on Dec 1, 2024 by Maddy Miller
In Technology with tags Explainer, Genetics
Part 2 in a series on Whole Genome Sequencing
1652 words, 6 minutes to read
This post is the second in a series about Whole Genome Sequencing, or WGS. The first article on choosing a whole genome sequencing provider and a quick primer on what it actually is can be found here.
If you’ve recently gotten a whole genome sequencing done and aren’t sure what to do with the giant files you’ve been given, this is a great place to start. This article goes into using a tool known as WGS Extract to convert between various formats, get some basic information about your genetic data, and export new data files to use with various platforms.
Downloading WGS Extract
WGS Extract can be found on their website, at the time of writing I’d recommend installing the v4 Beta to get the new features in the v4 release but without having to deal with the instability of the alpha or developer channels. If a more stable release channel of WGS Extract v4 is now available on the site, you should likely use that over the beta.
Installation of the program is as simple as extracting it, running the install script for your OS, and then running the WGSExtract script to run the program once it’s installed. For a more in-depth guide on installation, you can consult the WGS Extract manual.
Running WGS Extract
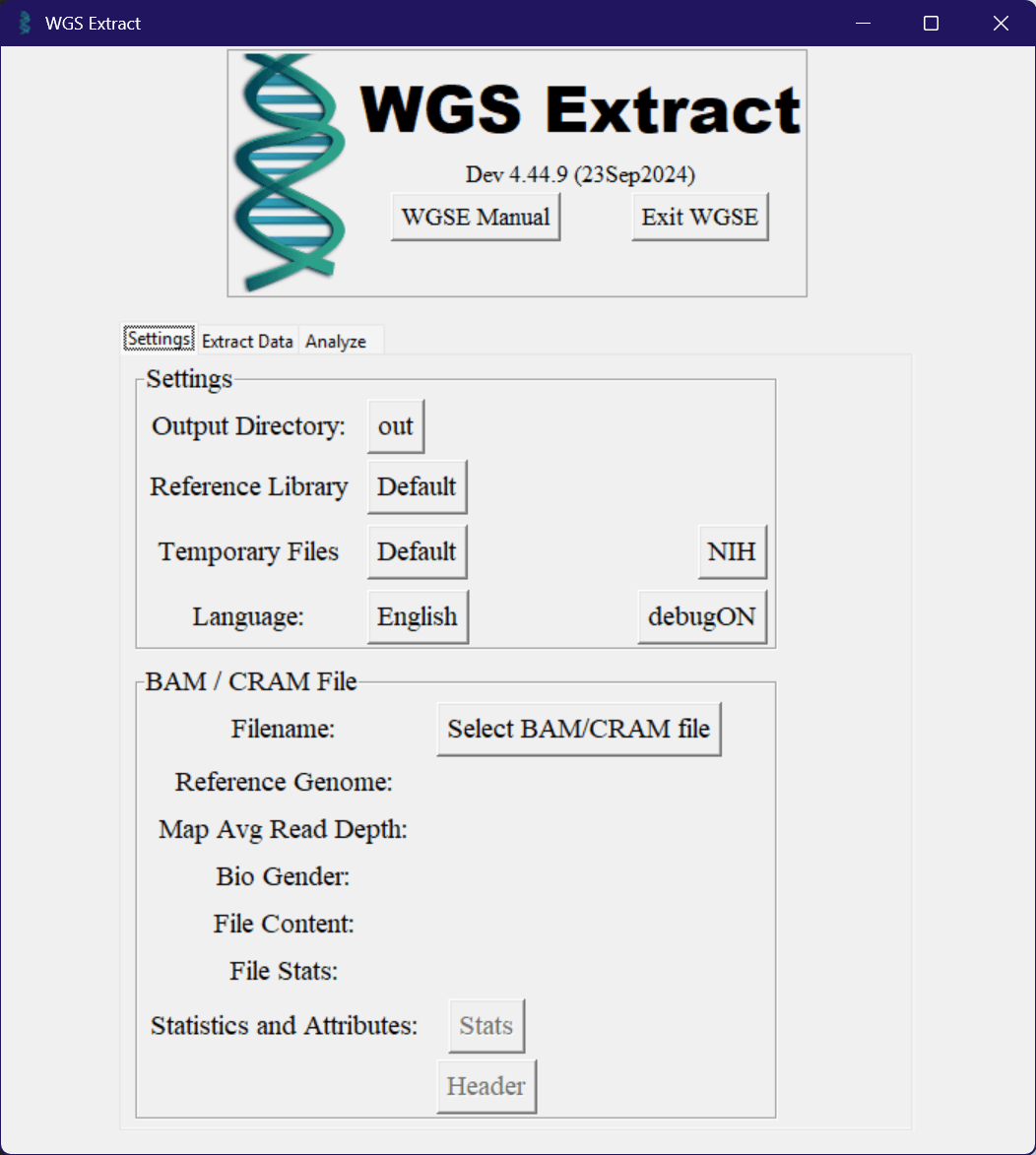
When you first open up the application, you should see something like this. The version you’re using will probably be different (hopefully newer), but other than that it should look mostly the same. First up, you want to make sure to select an output directory that WGS Extract will put any created or converted files into, using the button next to the “Output Directory” label in settings. Given the size of genetic data and the fact that temporary files might need to be created depending on the task you’re performing, it’s recommended to have a few hundred gigabytes of space available.
The next step you take is going to depend on what file type your genetic data is in. If you’ve got it in BAM or CRAM format you can use the “Select BAM/CRAM file” button to load the file, but if it’s something else it’ll require some level of conversion.
Starting from FASTQ files
If your genetic data came in the form of FASTQ files, you can use WGS Extract to create a new BAM file from your FASTQ files. To do this, click the “Analyze” tab within WGS Extract, and press the red “Align” button found in the “FASTQ file(s)” section.
This process might take a fairly long time, so it’s a good idea to start this when you’re able to leave your computer running for potentially hours depending on the speed of your computer.
What this information means
Ideally, if you've done everything correctly you should now have WGS Extract open with a BAM or CRAM file loaded with your genetic data. If everything has gone well, it should look something like this.
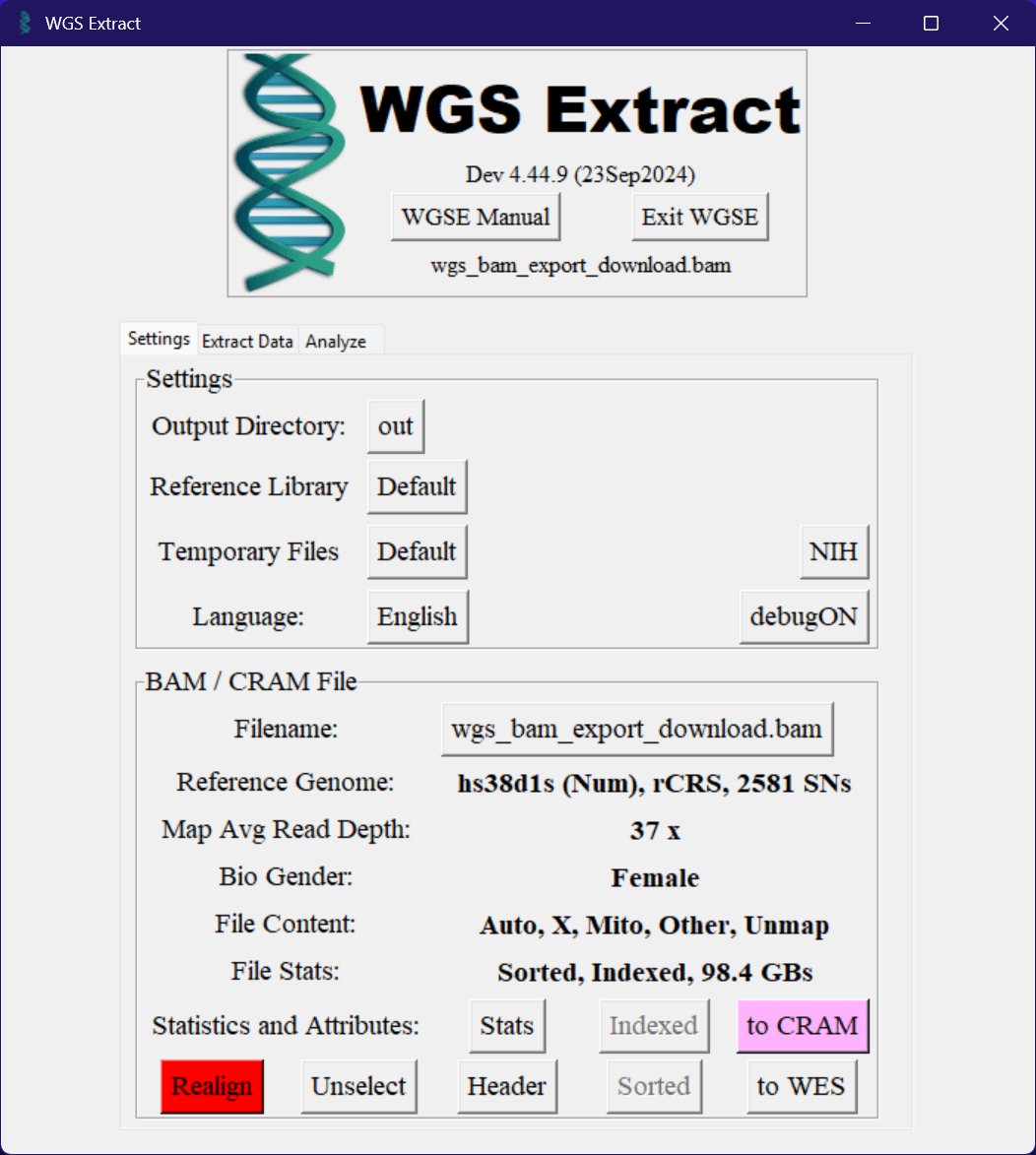
Here you can see some basic information about your genetic data, such as the reference genome used, the average read depth, the genetic sex of the sample, and what data is actually present in the file.
Reference Genome
As you can see from the above image of my sample, it was sequenced using the "hs38d1s" reference genome. For the purposes of the article this does not currently matter, but when attempting to use your genetic data directly with other platforms it's important to know what reference genome was used. Otherwise, the platform could be trying to do the equivalent of reading a book as if it's in English when it's actually written in French.
Average Read Depth
The next line shows the average read depth of the sample. With the way genetic sequencing is performed, it's not possible to perfectly read the whole thing precisely. So instead they perform a bunch of reads, up until the point where they should have read every part of the genome on average a certain number of times. The package I personally paid for was for 30x coverage, so the average read depth of my sample should be at least 30. This will vary between samples, even when they're sequenced at the same coverage level.
You can press the "Stats" button to get a more detailed breakdown of the average read depth for each individual chromosome. Generally this will be relatively close to the overall average read depth, with a notable exception being the mitochondrial DNA which will likely have a significantly higher read depth than the rest. Each cell contains a massive number of mitochondria, meaning the genome reading ends up covering it a very large number of times. The mitochondria isn't just the powerhouse of the cell, it's a whole industrial district.
File Content
The file content line shows what parts of the genome are present in the file. In the case of my sample, it has the autosomes (chromosomes 1-22), the X chromosome, and the mitochondrial DNA. If you're missing any of these, it's possible that the sequencing didn't work properly or that the data was lost at some point. Or alternatively, that the service you used didn't sequence those parts of the genome. If your DNA sample contains a Y chromosome, it should also be listed here.
Alongside these expected parts of the genome, you'll also notice there's an "Other" section and an "Unmapped" section. The "Other" section refers to any other DNA that was present in the sample that wasn't human, and the "Unmapped" section refers to any reads that didn't match against the reference genome. My sample was specifically of a buccal swab, so it will include the DNA of the various bacteria that was present in my mouth at the time. There are also a few viruses that latch themselves onto your DNA, which can also be present here. For example if you've ever been infected with the Epstein-Barr virus, it's possible that some of its DNA is still present in your cells and therefore the sequenced genome. The "Unmapped" section is generally very small, and is usually very low quality reads that couldn't be matched to the reference genome. Different reference genomes will have different amounts of unmapped reads, but in general they're not worth worrying about.
Haplogrouping
One of the functions that you can perform right from within WGS Extract, is determining your haplogroup. This is basically a code that refers to a specific branch of a human-wide family tree, finding common ancient ancestors. Others who share the same haplogroup with you aren't people you'd necessarily be related to in the way it's usually meant, but they are people you share a closer common ancestor with than someone who has a different haplogroup.
The haplogroup is generated from DNA which is considered very stable, meaning it doesn't change much between generations. This makes it a good tool to track ancestry over extremely long periods of time. As mitochondrial DNA is always passed exclusively from the mother, it can be used to trace your matrilineal ancestry, and as the Y chromosome is passed exclusively from the father it can be used to trace patrilineal ancestry if you have a Y chromosome. This does mean it has an inherent limitation in that it cannot trace your mother's father for example, or similar relationships higher up in the family tree.
If, like me, your DNA sample doesn't contain a Y chromosome, you won't be able to get your patrilineal haplogroup. If this is something you still want to find out, you can have either a genetically related brother (a half brother with a shared father will work) or your birth father submit a sample. Due to the stability of this DNA, their haplogroup would be the same as yours if you had a Y chromosome.
You can run a haplogroup from the "Analyze" tab, by pressing either the "Mitochondrial DNA" or "Y Chromosome" buttons. This will, after a few minutes or so, bring up a window like the one below.
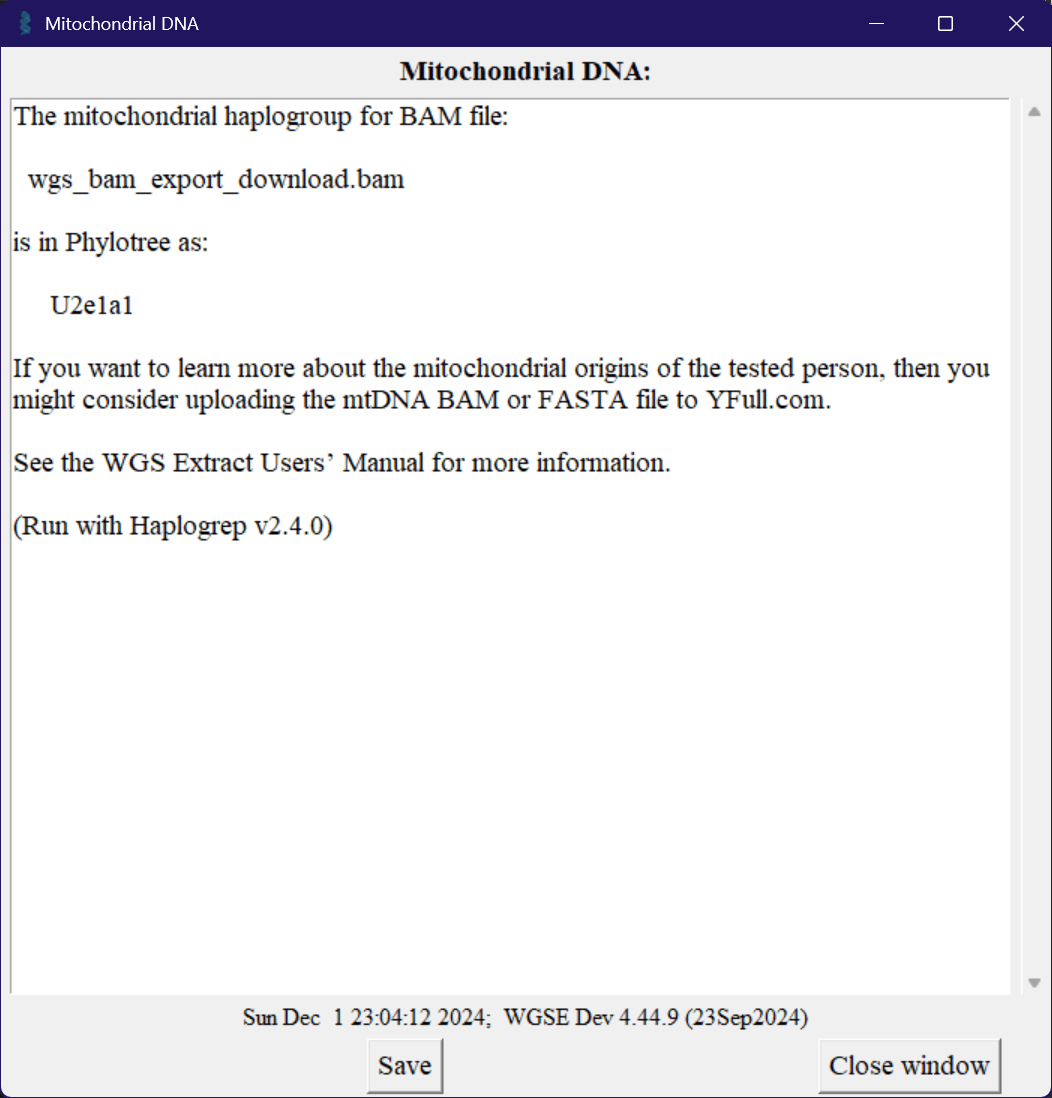
You can enter this haplogroup into various websites, such as YFull, or even just google it to see what discussions or research papers you can find. Because of how ancient these are, there are generally studies around population distribution and migration between continents of the ancestors of the haplogroup.
Exporting Data
If you want to upload your data to other platforms, you can use the "Extract Data" tab to convert to a few common formats. As can be seen in the image below, there are a few buttons available to output to a few different platforms.
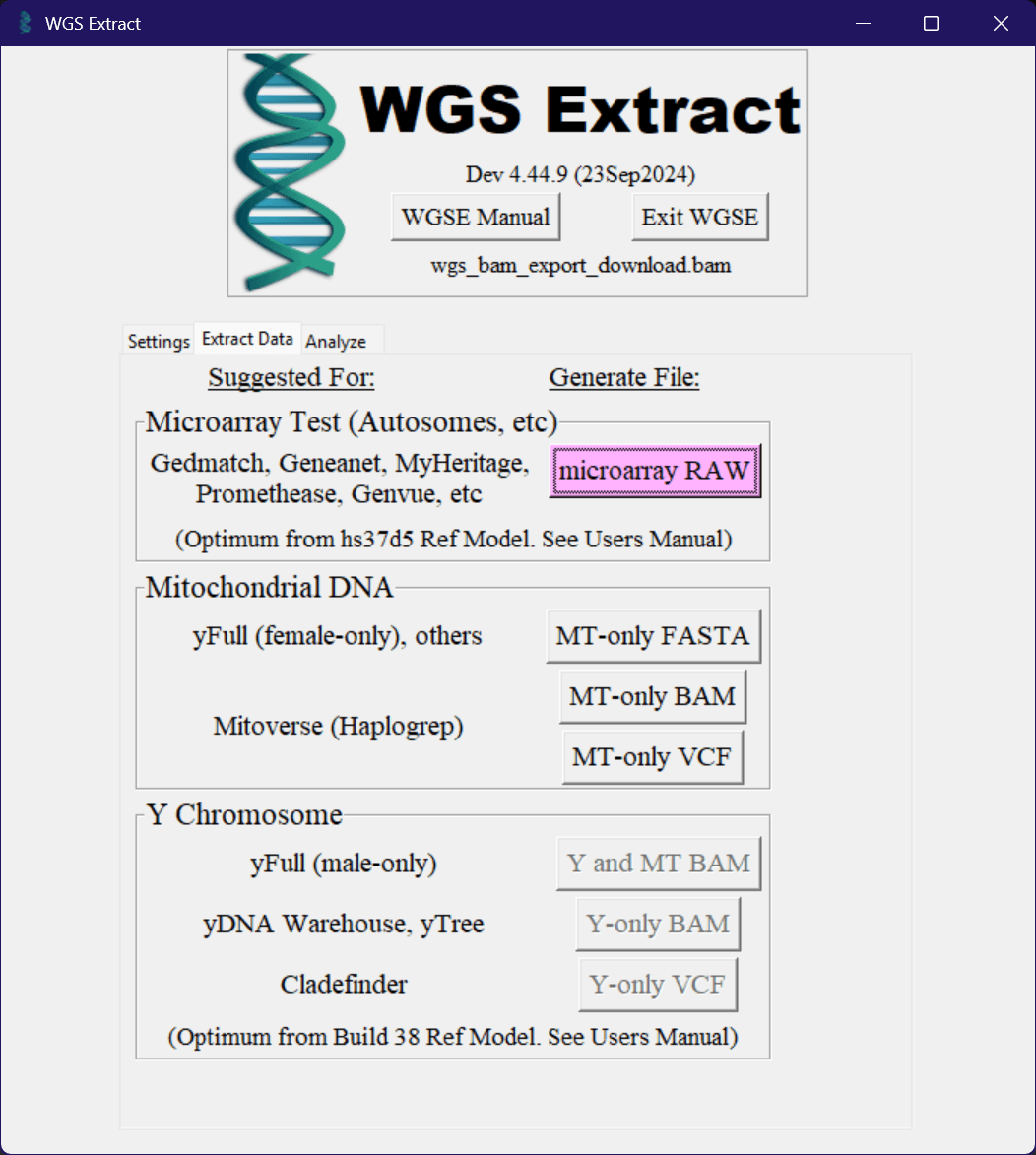
These buttons will convert your data to the format required by the platform, and then save it to the output directory you selected earlier, but can take a while to run depending on the speed of your computer.
Data Privacy and Risks
As I covered in the Data Privacy section of the last article, it's extremely important to be aware of what you're actually uploading to these platforms. While these exports are generally a fraction of the data of your WGS data, they still contain a lot of very sensitive information about you. While I understand this is why most people actually order a WGS, I'd urge you to understand the potential risks and implications of uploading your genetic data to these platforms. It's important to understand that these risks are not just for you, but also anyone related to you. If a health insurer got ahold of genetic data and used it to deny coverage or increase premiums, they could do so for anyone that you're related to as well.
Conclusion
WGS Extract is a powerful tool for converting between data formats, getting some basic statistics about your genetic data, and exporting data to other platforms. The next article on this topic covers ancestry analysis in more detail, including some local admixture tools that can partially emulate the online services without having to upload your data to third parties.
Continue reading the next part of the series: Whole Genome Sequencing Part 3: Ancestry Analysis



